[ad_1]
Summary: A new method using the gene-editing tool CRISPR-Cas9 has been developed to model liver cancer tumor subtypes caused by mutations in the same genes. By targeting a single section of the mouse gene, Ctnnb1, researchers were able to produce two distinct tumor subtypes, enhancing protein activity to promote tumor growth, which could allow for the development of new therapeutic interventions in the future.
Source: CSHL
Mutations in our genes can lead to severe problems, like colon or liver cancer. But cancer is very complex. Mutations in the same genes can lead to different subtypes of tumors in different people. Currently, scientists don’t have a good way to produce such tumor subtypes for study in the lab.
Now, Cold Spring Harbor Laboratory Assistant Professor Semir Beyaz has created a new method to model certain liver cancer tumor subtypes using the gene-editing tool CRISPR-Cas9.
Genes contain the information our bodies need to create proteins. Highly similar proteins produced from the same gene are called isoforms. Different isoforms generate different tumors. This process is known as exon skipping, where multiple parts of a gene are stitched together to make a different version of a protein.
“Everyone thinks that cancer is just one type,” Beyaz explains. “But with different isoforms, you can end up with cancer subtypes that have different characteristics.”
Beyaz and his colleagues produced two distinct tumor subtypes by targeting a single section of the mouse gene, Ctnnb1, with CRISPR. The tool is mostly used to inhibit gene function. This is the first time CRISPR has been used to generate different cancer-causing gain-of-function mutations in mice.
These mutations enhance protein activity to promote tumor growth. The team sequenced each tumor subtype to figure out which isoform was associated with the differences they observed.
“We were able to define those isoforms that associated with different cancer subtypes,” Beyaz says. “That was, for us, a surprising discovery.”
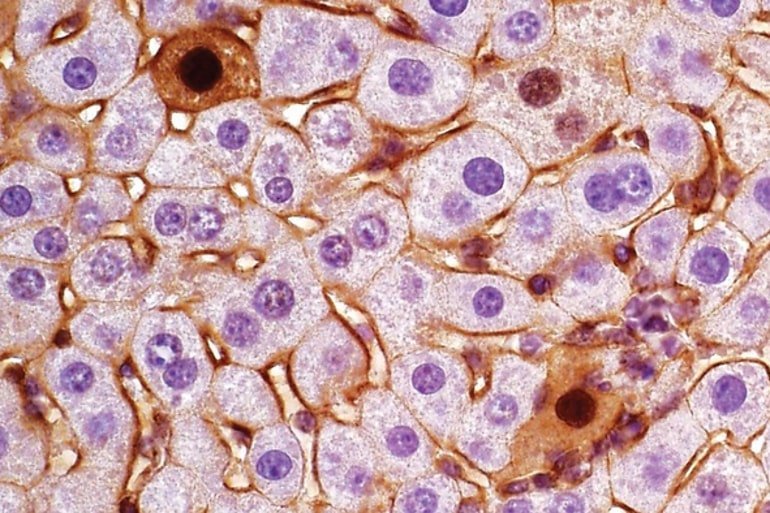
Next, to confirm that these isoforms actually caused the variances, they produced them in the mouse without using CRISPR. They found that they were indeed able to generate the two different tumor subtypes with their respective characteristics. Both of these liver tumor subtypes are also found in humans.
The mutations Beyaz targeted can lead to colon and liver cancers. Targeting exon skipping has emerged as a potential therapeutic approach for treating cancer and other diseases. Beyaz’s new study method allows researchers to investigate this phenomenon in living mice cells using CRISPR. The platform could someday help researchers develop new therapeutic interventions.
“Ultimately,” Beyaz explains, “what we want to do is find the best models to study the biology of cancer so that we can find a cure.”
Summary written with the assistance of ChatGPT AI technology
About this cancer and genetics research news
Author: Samuel Diamond
Source: CSHL
Contact: Samuel Diamond – CSHL
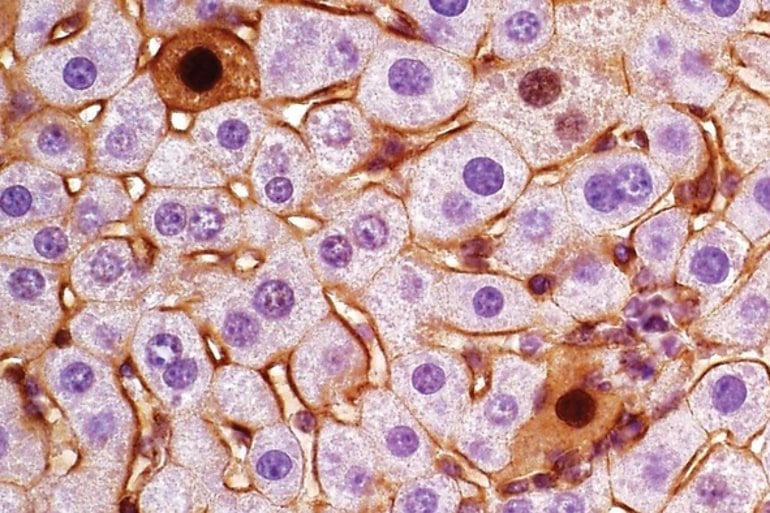
Image: The image is credited to Beyaz lab/Cold Spring Harbor Laboratory
Original Research: Open access.
“CRISPR-induced exon skipping of β-catenin reveals tumorigenic mutants driving distinct subtypes of liver cancer” by Semir Beyaz et al. Journal of Pathology
Abstract
CRISPR-induced exon skipping of β-catenin reveals tumorigenic mutants driving distinct subtypes of liver cancer
CRISPR/Cas9-driven cancer modeling studies are based on the disruption of tumor suppressor genes by small insertions or deletions (indels) that lead to frame-shift mutations.
In addition, CRISPR/Cas9 is widely used to define the significance of cancer oncogenes and genetic dependencies in loss-of-function studies. However, how CRISPR/Cas9 influences gain-of-function oncogenic mutations is elusive.
Here, we demonstrate that single guide RNA targeting exon 3 of Ctnnb1 (encoding β-catenin) results in exon skipping and generates gain-of-function isoforms in vivo. CRISPR/Cas9-mediated exon skipping of Ctnnb1 induces liver tumor formation in synergy with YAPS127A in mice. We define two distinct exon skipping-induced tumor subtypes with different histological and transcriptional features.
Notably, ectopic expression of two exon-skipped β-catenin transcript isoforms together with YAPS127A phenocopies the two distinct subtypes of liver cancer. Moreover, we identify similar CTNNB1 exon-skipping events in patients with hepatocellular carcinoma. C
ollectively, our findings advance our understanding of β-catenin-related tumorigenesis and reveal that CRISPR/Cas9 can be repurposed, in vivo, to study gain-of-function mutations of oncogenes in cancer.
[ad_2]
Source link